
Research

Overview
Species and ecosystems are inherently connected to the environments in which they evolve. The central theme of my lab’s research aims to understand the interplay between spatial, genetic, ecological and evolutionary processes. My lab integrates theoretical evolutionary and population genetic perspectives with geospatial, field-based and molecular genetic research to address fundamental questions regarding the evolution of molecules, organisms, populations, and ecosystems. We study the spatial patterns and processes driving community structure, distribution patterns, speciation, and biological diversity (genetic, phenotypic and behavioral). Most of the lab’s current research falls into two focal areas:
Conceptual foundations and methodological tools for quantifying the drivers of biodiversity patterns
Interplay between spatial, genetic and evolutionary processes
Understanding and predicting the spatio-temporal dynamics of populations remains a central goal in ecology. Much of my current research focuses on evaluating the genetic and demographic consequences of changes in habitat through time. The integration of geospatial and genetic data provide insights into the evolutionary mechanisms underlying recent divergences in ecology, phenotype and behavior (Brown & Knowles Mol. Ecol. 2012; Gehara, Summers & Brown 2013 Evo. Ecol.; French & Brown 2019 Mol. Phylo. Evo.) and the genetic consequences of future climate change (Brown et al. 2016 Am. J. Botany).
To understand spatio-temporal dynamics of genetics and demographics, we sample the genotypes of populations using sub-genomic data (e.g. in Pika, a Penstemen flower, and anole lizards). We then couple species distribution models (SDMs), paleogeographic data, and demographic models to emulate the colonization history of focal species. Demographic models are based on empirical population data (i.e., growth and migration rates, population densities). To specify unknown demographic parameters, Approximate Bayesian Computation is used to identify parameters that are biologically relevant and reflect our empirical genetic data. Following demographic modeling, coalescent genealogies are parameterized by the simulated demographic history. The genetic simulations match conditions that correspond to empirical genetic data sets (e.g., population number and location, number and length of loci). Using this framework, researchers can create explicit predictions of demographic and genetic changes across a landscape and can test specific hypotheses. These tests reject demographic histories that do not result in similar genetic values and marginal densities to the observed genetic data (Prates et al. 2017 PNAS).
Quantifying Niche Evolution
Understanding the drivers of the distributions of species, communities and ecosystems remains a fundamental aim across ecology and evolution. Within this topic, one broad field of study seeks to characterize and compare the ecological niches of species, with the ultimate goal of assessing how niches evolve. In the last few years, in collaboration with Ana Carnaval (CCNY), we have been developing novel quantitative tests of niche divergence in environmental space. These new quantitative methods address two major shortfalls of other methods that rely on assumptions of equilibrium distributions and are biased by the spatial abundance and availability of environments. These methods have been released as an open-source R package named humboldt (Brown & Carnaval 2019 Front. Biogeo).
We are also interested in quantifying the biogeographical consequences of niche evolution through time. Historical biogeographical methods typically quantify species’ ancestral distributions based purely on contemporary distributions of daughter species/lineages and often estimated ancestral distributions that are comprised of the distributions of all daughter species. In collaboration with my graduate student, Wilson Guillory, we are developing a novel method for estimating ancestral distributions based on quantifying the ecological tolerances of focal species and then estimating their evolution in a phylogenetic framework. These models are then projected into corresponding paleoclimatic data to characterize their geographic distributions at key time periods. Our method is incredibly unique because it allows for ancestral distributions to evolve in a manner that is not directly connected to the contemporary distributions of existing lineages, rather the ecological tolerances of contemporary species. Our methods will be released as an open-source freely available R package named machuruku to be used alongside a recently released paleoclimate database (www.paleoclim.org) created in collaboration with paleoclimatologists A. Haywood & D. Hill (U of Leeds).
Mechanisms of Biodiversity Patterns
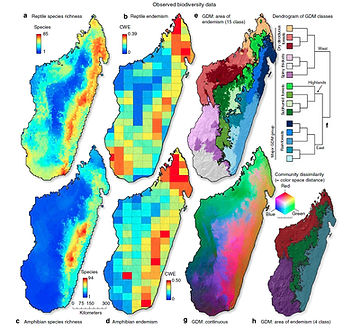
Another focus of my research has been to measure and model biodiversity patterns of large taxonomic groups and their underlying evolutionary and ecological processes. For example, in collaboration with M. Vences (Technical U Braunschweig) and A. Yoder (Duke U), I evaluated the spatial distribution patterns of Madagascar’s amphibians, reptiles and lemurs. Madagascar contains an extraordinary number of endemic flora and fauna. Given the incredible levels of endemism and the long period of isolation from other landmasses, this situation provides a unique opportunity to study the mechanisms driving divergence and diversification in situ. Furthermore, ongoing habitat destruction and climate change add urgency to studies of the flora and fauna of this remarkable and unique island country. The distribution of diverse organismal lineages will depend upon the idiosyncratic factors determined by their specific organismal life-histories combined with stochastic historical factors, and therefore, various assemblages of species are under the influence of differing mechanisms. (Brown et al. 2014 Nature Comm.; Brown et al. 2016 Plos One). Thus, any model that endeavors to explain island-wide patterns must necessarily be complex.
In collaboration with A. Carnaval, my research on this topic is extended to the incredibly diverse and threatened, Brazilian Atlantic Forest. We are studying how evolution within taxonomic groups shapes the broad-scale distribution of unique biological diversity across the landscape. In this research we move beyond traditional biodiversity measurements by quantifying and spatially modeling the phylogenetic diversity of six broad taxonomic groups spanning birds, plants to insects . Overall, my research has identified areas, or species, of high conservative priority, many which are currently being consider by authorities or have already resulted in additional protection (e.g., Madagascar rosewoods species were CITES designated globally; Barrett, Brown & Yoder 2010 Science, 2013 Nature). Many of our biodiversity quantification methods are released in a regularly updated and open-source, freely available ArcGIS package named SDMtoolbox (www.sdmtoolbox.org ).




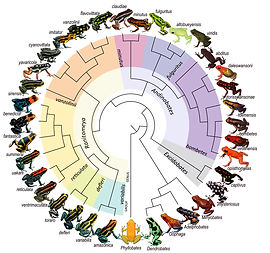

Diversification, speciation, and behavior of South American poison frogs
Neotropical poison frogs ignited my passion for research and they continue to drive a core tenant of my research program. They are a species-rich family (Dendrobatidae) that exhibit a diversity of mating systems, parental care strategies, and phenotypes. These tiny frogs often use their bright coloration to warn predators of their toxicity. In many cases, with increased female parental care, female choice drives color and patterns of populations to diverge. Due to these factors, these abundant diurnal frogs are an ideal model for studying parental care and color evolution, breeding ecology, and speciation in natural systems.
Biogeography, Phylogenetics and Systematics
I have a keen interest in the evolutionary histories of poison frogs. Several of my current projects focus on the poison frog genera: Ameerega and Ranitomeya, which recently diverged into the east Andes versant. We have already sequenced genomic data (UCEs) of over 850 tissue samples from all species in the genera from across the species’ ranges. I also have an extensive collection of acoustic and morphological data. These projects provide a rigorous framework for studying speciation, geographic variation – including widespread mimicry - and genome evolution. From a practical perspective, our systematic research clarifies the core evolutionary units, which gives meaning to the genes and genomic patterns we study and provides a starting point for biodiversity preservation. Lastly, my quantitative biogeography research provides an essential understanding of why species are in their present locations.
Environmental, spatial, and behavioral factors driving species boundaries
The key factors that reinforce species boundaries are poorly understood, particularly in natural settings. We currently are working with the Ameerega bassleri species complex, a group that possesses 3 species, each with distinct morphologies, distributions, and advertisement calls. All factors are thought to reinforce species boundaries in amphibians. Despite these distinctions, genomic data provide evidence of several pulses of historical hybridization between A. basssleri and the other two species in this complex. We are currently investigating the synergy between 1) the role of historic climate dynamism in these introgression events and 2) how the contemporary species phenotypes (acoustic and morphological) have maintained the mechanisms of speciation. To do the latter we are planning to use female choice experiments, which will allow us to distinguish the utility of acoustic and morphologically differences among species and their role in maintaining species boundaries.
Behavioral Ecology
The relationship between mating system and type of parental care is of fundamental importance to understanding the ecology and evolution of breeding systems. A large part of my behavioral research examines how the nature of reproductive resources used by species influences the reproductive strategies of individuals, including the mating system, the form of parental care, and levels of sexual conflict. In Peru, I worked with two sympatric species of closely related poison frogs that differ in parental care strategies. In collaboration with K. Summers (ECU), our results revealed that the transition to rearing tadpoles in tiny pools (mediated by competition and sexual conflict) drove the evolution of biparental care and monogamy. We also demonstrated the first case of brood parasitism (Brown et al. 2008 Biol. Lett.) and genetic monogamy in anurans (Brown et al. 2010 Am. Nat.).
